Access of Data
| Data | URL |
|---|---|
| image | gs://neuroglancer/zfish_v1/image |
| segmentation | gs://neuroglancer/zfish_v1/consensus-20190415 |
| cleft | gs://neuroglancer/zfish_v1/clefts |
| skeletons | https://drive.google.com/drive/folders/1UEb398-fekzqHrhv8JAgszW99Na9aGlK?usp=sharing |
| synapses | https://drive.google.com/file/d/19R-C1uUHQqwhq6j65AiKk-kWlYkEeaKM/view?usp=sharing |
Visualization
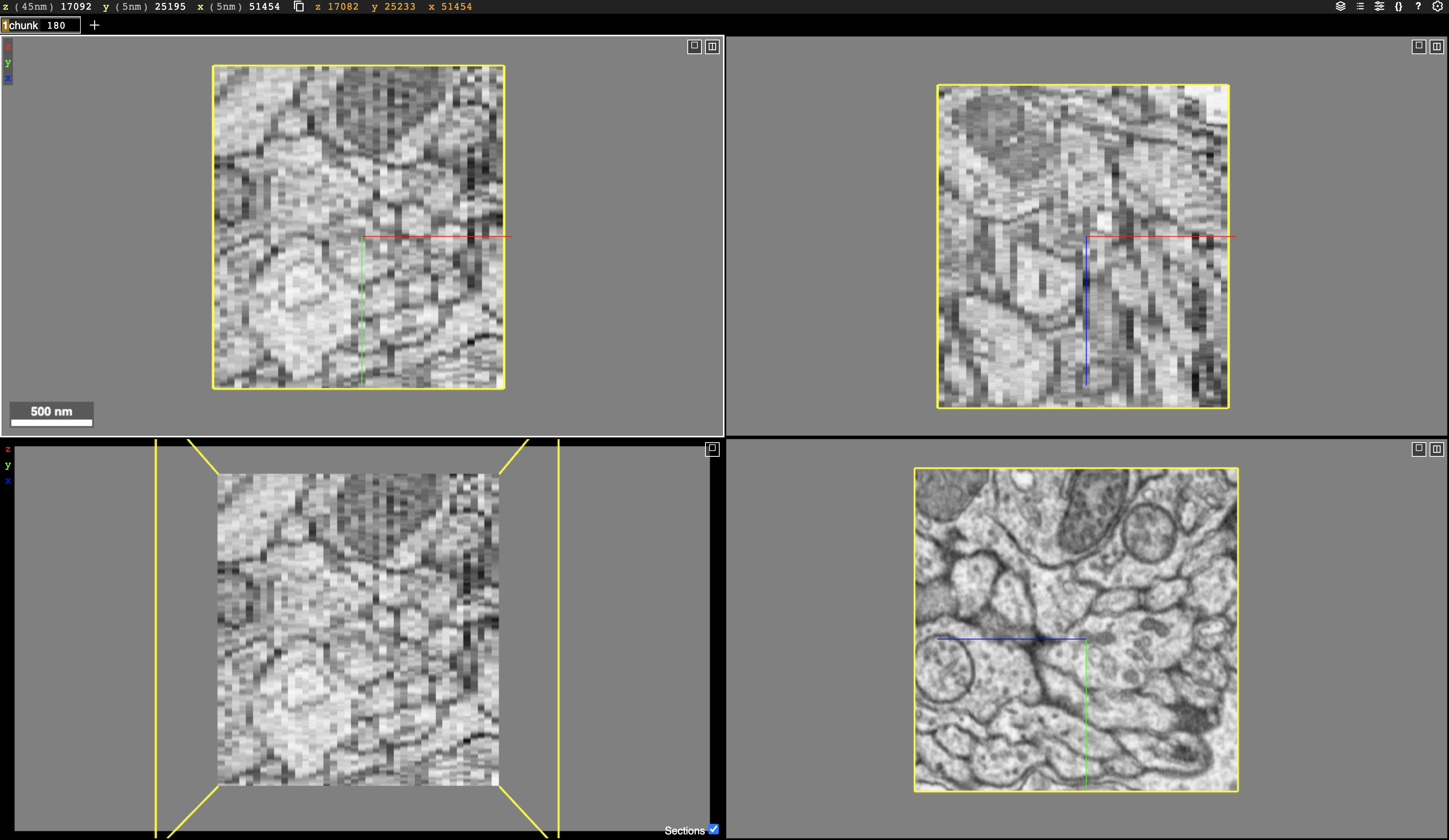
We use Neuroglancer to visualize the image, segmentation, skeleton, and meshes. This is an example link containing image and segmentation (sparsely proofread). More example links are in the Gallery page. Synapses can also be visualized using Neuroglancer with some customized code.
Navigation
Here is a link describing how to navigate once you are on the neuroglancer page link. The Seung lab fork of neuroglancer can be found here link.
Downloading
For Google Drive links, the data could be downloaded directly by clicking the links.
Image and Segmentation
Image and segmentation volumes are stored in Google Cloud Storage. You can cutout some chunks using CloudVolume or chunkflow(still using CloudVolume under the hood). For whole dataset downloading, it is recommended to contact us first.
NOTE: CloudVolume use f-order (XYZ) while chunkflow use c-order (ZYX) in default!
NOTE: https should be used to avoid using credential files.
Example code to cutout an image chunk using CloudVolume:
from cloudvolume import CloudVolume
vol = CloudVolume('precomputed://gs://neuroglancer/zfish_v1/image', use_https=True)
img = vol[51267:51367, 24985:25085, 17072:17172]
Example commandline to cutout an image chunk using chunkflow. This command also saves the image chunk to local disk as a HDF5 file and visualize it using Neuroglancer.
chunkflow load-precomputed -v precomputed://gs://neuroglancer/zfish_v1/image --use-https --chunk-start 17072 24985 51267 --chunk-size 40 400 400 save-h5 -f image.h5 neuroglancer
After that you should see a link in the terminal. Click the link, you should see the image volume visualized in webbrowser.
NOTE: If the link is something like http://c02cq0glmd6v.fios-router.home:55676/v/29bd625ab5c364d68ad9f9f37b58a5444d64de56/, try to replace the
c02cq0glmd6v.fios-router.homewithlocalhost.
Meshes
You can visualize the meshes by clicking the objects in Neuroglancer.
In order to download the meshes, you can use CloudVolume.
The following is an example code modified from CloudVolume documentation:
from cloudvolume import CloudVolume
vol = CloudVolume('gs://mylab/mouse/image', parallel=True, progress=True)
label = 1
mesh = vol.mesh.get(label)
vol.mesh.save(12345) # save 12345 as ./12345.ply on disk
vol.mesh.save([12345, 12346, 12347]) # merge three segments into one file
vol.mesh.save(12345, file_format='obj') # 'ply' and 'obj' are both supported
vol.mesh.get(12345) # return the mesh as vertices and faces instead of writing to disk
vol.mesh.get([ 12345, 12346 ]) # return these two segids fused into a single mesh
vol.mesh.get([ 12345, 12346 ], fuse=False) # return { 12345: mesh, 12346: mesh }
vol.mesh.put(meshes) # works for unsharded legacy only
vol.mesh.delete(segids) # works for unsharded meshes only
mesh.viewer() # Opens GUI. Requires vtk.
Screenshots and Video Recordings
Neuroglancer provides internal tools for screenshots and video recording. The documentation of usage is embedded in the code.
Example usages is as follows:
python -m neuroglancer.tool.screenshot --help
python -m neuroglancer.tool.video_tool --help
python -m neuroglancer.tool.screenshot state.json
python -m neuroglancer.tool.video_tool script.txt